1. Ubiquitin and proteasome system proteolysis
Lysosomal proteolysis, calpain, and proteasome proteolysis through ubiquitin are known as the main systems for intracellular proteolysis.
Particularly, expression regulation through ubiquitin interstitial proteolysis has the following features: 1. Irreversible biochemical reactions; and 2. It is a system that proteolyzes the target protein quickly.
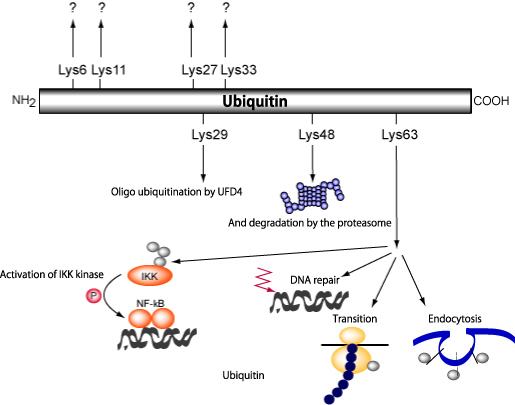
Figure 1: High order intracellular functions controlled by ubiquitin/
proteasome system control.
Ubiquitination is related to many intracellular phenomena in terms of function; it is particularly involved in intercellular signaling, cell cycles, membrane receptor interstitial endocytosis, transcription, organellar composition, spermatogenesis modification, etc.
Research into the biological reactions of ubiquitin interstitial proteasome proteolysis systems has seen remarkable progress in recent times, ubiquitin activating enzyme (E1), ubiquitin-ligase (E2), ubiquitin-protein ligase (E3) have been molecularly cloned, and the enzymes involved in the specific ubiquitination of various target proteins have been identified.
Protein that have been the subject of ubiquitination are recognized as molecules called Rpn10/S5a that exist in the 19S proteasome cap, and undergo degradation through the a7b7 subunit that makes up 20S.
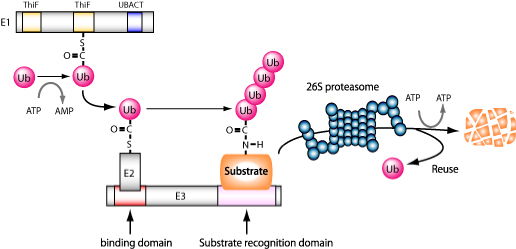
Figure 2: Ubiquitination reaction
Ubiquitination reactions are in the form of post-translational modifications of proteins catalyzed by cascades composed of E1 (ubiquitin activating enzyme), E2(ubiquitin-conjugating enzyme), and E3 (ubiquitin-igase). Target proteins that have undergone polyubiquitination are recognized as proteasomes and degraded.
2. The diversity of ubiquitin-ligase E3
As part of the progress in the analysis of ubiquitination on a molecular level, the ubiquitin-ligase, called E6AP(E6-associated protein), which acts as the E3 that binds with the human papillomavirus gene product and degrades the tumor suppressor gene p53 has been identified.
With regard to important cancer suppressor gene products 53, which are transcription factors that monitor damage to DNA, among the cells that have been infected with human papillomavirus (particularly cervical squamous atypical cells), E6, which is one of these gene products, physically takes on a trimeric form by binding to p53 and E6AP and it is reported that the E6AP, which has ubiquitin-ligase activation properties, exhibits the ubiquitination of p53.
E6AP has ubiquitin and cysteine residue, which activates thioester bond, on its C-terminal side; this discovery is evidence that ubiquitination exists on a molecular level as a E1-E2-E3 ubiquitination transfer reaction
Further, the approximately 300 amino acids that comprise these ubiquitin binding sites are known to be widely preserved in arrangements of numerous proteins in various eukaryotic cells; called HECT domain (homologous to E6AP carboxy terminus), it is recognized that proteins possessing this domain function as ubiquitin-ligases.
Currently, aside from E6AP, Nedd4、Rsp5、Pub1、Tom1、Ufd4、Smurf1/2、p100 have also been identified.
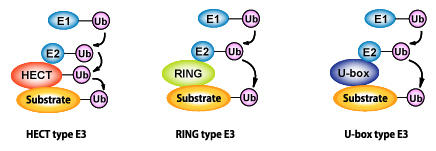
Figure 3: Diverse ubiquitin-ligases (E3)
Ubiquitin-ligase (E3) is an enzyme that controls the substrate recognition of ubiquitination, and, currently, is classified into three ubiquitin-ligase groups based on differences in domain structure, they are: HECT type, RING type, and U-box type. HECT type E3 itself binds with cysteine residue to form an intermediate that forms thioester bonds with ubiquitin.
It has been indicated that Cbl is involved in the stability of PDGF and EGF receptors. In actuality, Cbl has, before, been called the RING finger domain (really interesting new gene) and is a module that binds with zinc, and it is that portion that is involved in the stability of a subject protein.
At the same time, through the screening of yeast two-hybrid of human E2, called UbcH5B, which has been made the bait, a molecule possessing a RING finger (AO7) has been identified; and, the recognition of the ubiquitin activating properties of molecules with the RING finger domain has been confirmed.
The SCF(Skp1/Cul1/F-box protein) complex has been identified (SCFFbw1) as the ubiquitin-ligase molecular complex that controls the interstitial ubiquitin degradation of IkB, an NF-kB signal inhibitory system regulatory molecule, and the interstitial molecule β-catenin of the Wnt/Wingless signal; after this, SCFSkp2 has been identified as the ubiquitin-ligase that has effects on CDK inhibitor p27, important to transition at the GS/S stage of the cell cycle, as well as the ubiquitination of cyclin E.
Further, Rbx1/ROC1 has been identified as the 4th structural component of SCF, and it has been shown that there is a RING finger motif present in this molecule.
Also, the composite molecules of APC/C (anaphase promoting complex/ cyclosome), which performs the ubiquitination of M-phase cyclin cyclin B, have been comprehensively identified; and it has been shown that APC11, which possesses the RING finger domain, is present in it.
The above two RING finger proteins manifest their ubiquitin-activation properties as complexes, however, there also exist other molecules that act as ubiquitin-ligases even when alone, such as the aforementioned Cbl, and also Mdm2、IAP、Siah-1、BRCA1, etc.
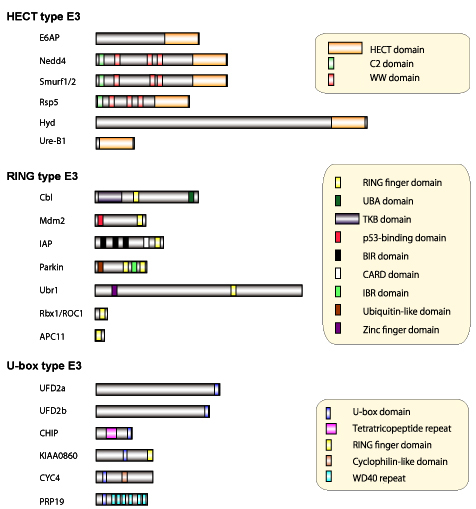
Figure 4: Molecular structure of diverse ubiquitin-ligases (E3)
Screening is conducted using the genetics of budding yeast to identify the molecules that control the degradation of the fusion protein (Ub-bgal) of ubiquitin and b-galactosidase, using their stabilities as indicators.
It has been reported that a ubiquitin reaction system can be achieved through the use of a molecule that has been shown to increase the length of the chains of E1、E2(Ubc4) and UFD4 (HECT type ubiquitin ligase), and a polyubiquitin called UFD2.
Though structural analysis, it has been shown that the regions where the HECT domain and the RING domain bind with E2 resemble each other; and, through computer database analysis, it is clear that the area called the "U-box" of this UFD2, in terms of the arrangement of proteins, resembles the RING domain.
- Department of Biochemistry,
Faculty of Medicine and Graduate School of Medicine,
Hokkaido University - Kita 15, Nishi 7, Kita-ku, Sapporo,
Hokkaido 060-8638, Japan
TEL: +81-11-706-5047
FAX: +81-11-706-5169